armi.nucDirectory.nuclideBases module
This module provides access to fundamental nuclide information to be used throughout the framework and applications.
Implementation: Isotopes and isomers can be queried by name, label, MC2-3 ID, MCNP ID, and AAAZZZS ID. I_ARMI_ND_ISOTOPES0
|
The The The
|
The nuclide class structure is outlined here.
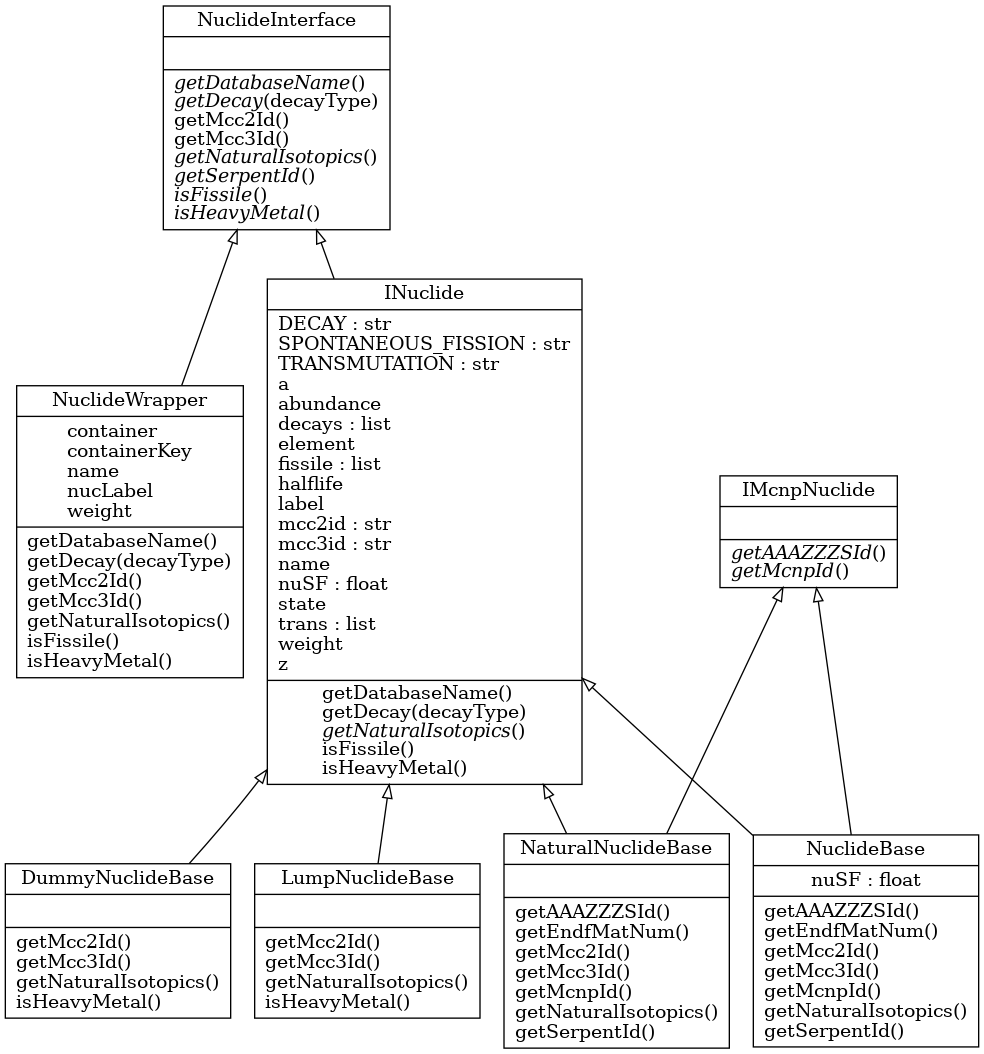
Class inheritance diagram for
INuclide.
Examples
>>> r = Reactor("ExampleReactor", bp)
>>> r.nuclideBases.byName["U235"]
<NuclideBase U235: Z:92, A:235, S:0, W:2.350439e+02, Label:U235>, HL:2.22160758861e+16, Abund:7.204000e-03>
>>> r.nuclideBases.byLabel["U235"]
<NuclideBase U235: Z:92, A:235, S:0, W:2.350439e+02, Label:U235>, HL:2.22160758861e+16, Abund:7.204000e-03>
Retrieve U-235 by the MC2-2 ID:
>>> r.nuclideBases.byMcc2Id["U-2355"]
<NuclideBase U235: Z:92, A:235, S:0, W:2.350439e+02, Label:U235>, HL:2.22160758861e+16, Abund:7.204000e-03>
Retrieve U-235 by the MC2-3 ID:
>>> r.nuclideBases.byMcc3IdEndfVII0["U235_7"]
<NuclideBase U235: Z:92, A:235, S:0, W:2.350439e+02, Label:U235>, HL:2.22160758861e+16, Abund:7.204000e-03>
Retrieve U-235 by the MCNP ID:
>>> r.nuclideBases.byMcnpId["92235"]
<NuclideBase U235: Z:92, A:235, S:0, W:2.350439e+02, Label:U235>, HL:2.22160758861e+16, Abund:7.204000e-03>
Retrieve U-235 by the AAAZZZS ID:
>>> r.nuclideBases.byAAAZZZSId["2350920"]
<NuclideBase U235: Z:92, A:235, S:0, W:2.350439e+02, Label:U235>, HL:2.22160758861e+16, Abund:7.204000e-03>
Notes
Currently, this module contains a lot of data in the global scope. But ARMI is in the process of encapsulating this data, moving it out of the global scope, making it part of the reactor data model, and making it configurable via Settings. Pardon the mess during this transition.
- class armi.nucDirectory.nuclideBases.NuclideInterface[source]
Bases:
objectAn abstract nuclide implementation which defining various methods required for a nuclide object.
- getDecay(decayType)[source]
Return a
DecayModeobject.- Parameters:
decType (str) – Name of decay mode, e.g. ‘sf’, ‘alpha’
- Returns:
decay
- Return type:
- getMcc2Id()[source]
Return the MC2-2 nuclide identification label based on the ENDF/B-V.2 cross section library.
- getMcc3Id()[source]
Return the MC2-3 nuclide identification label based on the ENDF/B-VII.1 cross section library.
- getMcc3IdEndfbVII0()[source]
Return the MC2-3 nuclide identification label based on the ENDF/B-VII.0 cross section library.
- class armi.nucDirectory.nuclideBases.NuclideWrapper(container, key)[source]
Bases:
NuclideInterfaceA nuclide wrapper class, used as a base class for nuclear data file nuclides.
- property name
Return the underlying nuclide’s name (i.e. “PU239”).
Notes
The nuclide name consists of the capitalized 2 character element symbol and atomic mass number.
- property weight
Get the underlying nuclide’s weight.
- getDecay(decayType)[source]
Return a
DecayModeobject.- Parameters:
decType (str) – Name of decay mode, e.g. ‘sf’, ‘alpha’
- Returns:
decay
- Return type:
- getMcc3IdEndfbVII0()[source]
Return the MC2-3 nuclide based on the ENDF/B-VII.0 cross section library.
- class armi.nucDirectory.nuclideBases.INuclide(element, a, state, weight, abundance, halflife, name, label, mcc2id=None, mcc3idEndfbVII0=None, mcc3idEndfbVII1=None)[source]
Bases:
NuclideInterfaceNuclide interface, the base of all nuclide objects.
- Variables:
z (int) – Number of protons.
a (int) – Number of nucleons.
state (int) – Indicates excitement, 1 is more excited than 0.
abundance (float) – Isotopic fraction of a naturally occurring nuclide. The sum of all nuclide abundances for a naturally occurring element should be 1.0. This is atom fraction, not mass fraction.
name (str) – ARMI’s unique name for the given nuclide.
label (str) – ARMI’s unique 4 character label for the nuclide. These are not human readable, but do not lose any information. The label is effectively the
Element.symbolpadded to two characters, plus the mass number (A) in base-26 (0-9, A-Z). Additional support for meta-states is provided by adding 100 * the state to the mass number (A).nuSF (float) – Neutrons released per spontaneous fission. This should probably be moved at some point.
Create an instance of an INuclide.
Warning
Do not call this constructor directly; use the factory instead.
- fissile = ['U235', 'PU239', 'PU241', 'AM242M', 'CM244', 'U233']
- TRANSMUTATION = 'transmutation'
- DECAY = 'decay'
- SPONTANEOUS_FISSION = 'nuSF'
- getDecay(decayType)[source]
Get a
DecayMode.Retrieve the first
DecayModematching the specified decType.- Parameters:
decType (str) – Name of decay mode e.g. ‘sf’, ‘alpha’
- Returns:
decay
- Return type:
- class armi.nucDirectory.nuclideBases.IMcnpNuclide[source]
Bases:
objectAbstract class for retrieving nuclide identifiers for the MCNP software.
- class armi.nucDirectory.nuclideBases.NuclideBase(element, a, weight, abundance, state, halflife)[source]
Bases:
INuclide,IMcnpNuclideRepresents an individual nuclide/isotope.
The
NuclideBaseclass provides a data structure for information about a single nuclide, including the atom number, atomic weight, element, isomeric state, half-life, and name. The class contains static methods for creating an internal ARMI name or label for a nuclide. There are instance methods for generating the nuclide ID for external codes, e.g. MCNP or Serpent, and retrieving the nuclide ID for MC2-2 or MC2-3. There are also instance methods for generating an AAAZZZS ID and an ENDF MAT number.- getNaturalIsotopics()[source]
Gets the natural isotopics root
Element.Gets the naturally occurring nuclides for this nuclide.
See also
- getMcc2Id()[source]
Return the MC2-2 nuclide identification label based on the ENDF/B-V.2 cross section library.
This method returns the
mcc2idattribute of aNuclideBaseinstance. This attribute is initially populated by reading from the mcc-nuclides.yaml file in the ARMI resources folder.
- getMcc3Id()[source]
Return the MC2-3 nuclide identification label based on the ENDF/B-VII.1 cross section library.
- getMcc3IdEndfbVII0()[source]
Return the MC2-3 nuclide identification label based on the ENDF/B-VII.0 cross section library.
This method returns the
mcc3idEndfbVII0attribute of aNuclideBaseinstance. This attribute is initially populated by reading from the mcc-nuclides.yaml file in the ARMI resources folder.
- getMcc3IdEndfbVII1()[source]
Return the MC2-3 nuclide identification label based on the ENDF/B-VII.1 cross section library.
This method returns the
mcc3idEndfbVII1attribute of aNuclideBaseinstance. This attribute is initially populated by reading from the mcc-nuclides.yaml file in the ARMI resources folder.
- getMcnpId()[source]
Gets the MCNP label for this nuclide.
This method generates the MCNP ID for an isotope using the standard MCNP format based on the atomic number A, number of protons Z, and excited state. The implementation includes the special rule for Am-242m, which is 95242. 95642 is used for the less common ground state Am-242.
- Returns:
id – The MCNP ID e.g.
92235,94239,6000- Return type:
- getAAAZZZSId()[source]
Return a string that is ordered by the mass number, A, the atomic number, Z, and the isomeric state, S.
This method generates the AAAZZZS format ID for an isotope. Where AAA is the mass number, ZZZ is the atomic number, and S is the isomeric state. This is a general format independent of any code that precisely defines an isotope or isomer.
Notes
An example would be for U235, where A=235, Z=92, and S=0, returning
2350920.
- getSerpentId()[source]
Returns the SERPENT style ID for this nuclide.
- Returns:
id – The ID of this nuclide based on it’s elemental name, weight, and state, eg
U-235,Te-129m.- Return type:
- getEndfMatNum()[source]
Gets the ENDF MAT number.
MAT numbers are defined as described in section 0.4.1 of the NJOY manual. Basically, it’s Z * 100 + I where I is an isotope number. I=25 is defined as the lightest known stable isotope of element Z, so for Uranium, Z=92 and I=25 refers to U234. The values of I go up by 3 for each mass number, so U235 is 9228. This leaves room for three isomeric states of each nuclide.
- Returns:
id – The MAT number e.g.
9237for U238- Return type:
- class armi.nucDirectory.nuclideBases.NaturalNuclideBase(name, element)[source]
Bases:
INuclide,IMcnpNuclideRepresents an individual nuclide/isotope that is naturally occurring.
Notes
This is meant to represent the combination of all naturally occurring nuclides within an element. The abundance is forced to zero here so that it does not have any interactions with the NuclideBase objects.
- getNaturalIsotopics()[source]
Gets the natural isotopics root
Element.Gets the naturally occurring nuclides for this nuclide.
See also
- getMcnpId()[source]
Gets the MCNP ID for this element.
- Returns:
id – The MCNP ID e.g.
1000,92000. Not zero-padded on the left.- Return type:
- getMcc2Id()[source]
Return the MC2-2 nuclide identification label based on the ENDF/B-V.2 cross section library.
- getMcc3Id()[source]
Return the MC2-3 nuclide identification label based on the ENDF/B-VII.1 cross section library.
- getMcc3IdEndfbVII0()[source]
Return the MC2-3 nuclide identification label based on the ENDF/B-VII.0 cross section library.
- getMcc3IdEndfbVII1()[source]
Return the MC2-3 nuclide identification label based on the ENDF/B-VII.1 cross section library.
- class armi.nucDirectory.nuclideBases.DummyNuclideBase(element, name, weight)[source]
Bases:
INuclideRepresents a dummy/placeholder nuclide within the system.
Notes
This may be used to store mass from a depletion calculation, specifically in the instances where the burn chain is truncated.
- getNaturalIsotopics()[source]
Gets the natural isotopics, an empty iterator.
Gets the naturally occurring nuclides for this nuclide.
- Returns:
empty – An empty generator
- Return type:
iterator
See also
- getMcc2Id()[source]
Return the MC2-2 nuclide identification label based on the ENDF/B-V.2 cross section library.
- getMcc3Id()[source]
Return the MC2-3 nuclide identification label based on the ENDF/B-VII.1 cross section library.
- class armi.nucDirectory.nuclideBases.LumpNuclideBase(element, name, weight)[source]
Bases:
INuclideRepresents a combination of many nuclides from NuclideBases into a single lumped nuclide.
See also
armi.physics.neutronics.fissionProductDescribes what nuclides LumpNuclideBase is expend to.
- getNaturalIsotopics()[source]
Gets the natural isotopics, an empty iterator.
Gets the naturally occurring nuclides for this nuclide.
- Returns:
empty – An empty generator
- Return type:
iterator
See also
- getMcc2Id()[source]
Return the MC2-2 nuclide identification label based on the ENDF/B-V.2 cross section library.
- getMcc3Id()[source]
Return the MC2-3 nuclide identification label based on the ENDF/B-VII.1 cross section library.
- armi.nucDirectory.nuclideBases.initReachableActiveNuclidesThroughBurnChain(nuclides, numberDensities, activeNuclides)[source]
Pass through to NuclideBases.initReachableActiveNuclidesThroughBurnChain() for the global NuclideBases object.
- armi.nucDirectory.nuclideBases.getIsotopics(nucName)[source]
Pass through to NuclideBases.getIsotopics() for the global NuclideBases object.
- armi.nucDirectory.nuclideBases.fromName(name)[source]
Pass through to NuclideBases.fromName() for the global NuclideBases object.
- armi.nucDirectory.nuclideBases.isMonoIsotopicElement(name)[source]
Pass through to NuclideBases.isMonoIsotopicElement() for the global NuclideBases object.
- armi.nucDirectory.nuclideBases.where(predicate)[source]
Pass through to NuclideBases.where() for the global NuclideBases object.
- armi.nucDirectory.nuclideBases.single(predicate)[source]
Pass through to NuclideBases.single() for the global NuclideBases object.
- armi.nucDirectory.nuclideBases.changeLabel(nuclideBase, newLabel)[source]
Pass through to NuclideBases.changeLabel() for the global NuclideBases object.
- armi.nucDirectory.nuclideBases.getDepletableNuclides(activeNuclides, obj)[source]
Get nuclides in this object that are in the burn chain.
- armi.nucDirectory.nuclideBases.imposeBurnChain(burnChainStream)[source]
Pass through to NuclideBases.imposeBurnChain() for the global NuclideBases object.
- armi.nucDirectory.nuclideBases.factory()[source]
Pass through to NuclideBases.factory() for the global NuclideBases object.
- armi.nucDirectory.nuclideBases.addNuclideBases()[source]
Pass through to NuclideBases.addNuclideBases() for the global NuclideBases object.
- armi.nucDirectory.nuclideBases.readMCCNuclideData()[source]
Pass through to NuclideBases.readMCCNuclideData() for the global NuclideBases object.
- armi.nucDirectory.nuclideBases.updateNuclideBasesForSpecialCases()[source]
Pass through to NuclideBases.updateNuclideBasesForSpecialCases() for the global NuclideBases object.
- armi.nucDirectory.nuclideBases.addGlobalNuclide(nuclide: NuclideBase)[source]
Pass through to NuclideBases.addNuclide() for the global NuclideBases object.
- armi.nucDirectory.nuclideBases.destroyGlobalNuclides()[source]
Pass through to NuclideBases.clear() for the global NuclideBases object.
- class armi.nucDirectory.nuclideBases.NuclideBases(nuclidesFile=None, mccNuclidesFile=None)[source]
Bases:
objectA container for all the nuclide information in the simulation.
By design, you would only expect to have one instance of this object in memory during a simulation.
- Variables:
burnChainImposed (bool) – Have we applied transmutation and decay data to each nuclide?
instances (list[INuclide]) – A simple list of the nuclides in this class.
byName (dict[str, INuclide]) – A dictionary of the nuclides in this class, keyed by name, e.g., “U235”.
byDBName (dict[str, INuclide]) – A dictionary of the nuclides in this class, keyed by database name, e.g., “nU235”.
byLabel (dict[str, INuclide]) – A dictionary of the nuclides in this class, keyed by label, e.g., “U235”.
byMcc2Id (dict[str, INuclide]) – A dictionary of the nuclides in this class, keyed by MC2-2 ID, e.g., “U-2355”.
byMcc3Id (dict[str, INuclide]) – A dictionary of the nuclides in this class, keyed by MC2-3 ID, e.g., “U235_7”. (This exists for backwards compat. Identical to byMcc3IdEndfbVII1.)
byMcc3IdEndfbVII0 (dict[str, INuclide]) – A dictionary of the nuclides in this class, keyed by MC2-3 ID, e.g., “U235_7”.
byMcc3IdEndfbVII1 (dict[str, INuclide]) – A dictionary of the nuclides in this class, keyed by MC2-3 ID, e.g., “U235_7”.
byMcnpId (dict[str, INuclide]) – A dictionary of the nuclides in this class, keyed by MCNP ID, e.g., 92235.
byAAAZZZSId (dict[int, INuclide]) – A dictionary of the nuclides in this class, keyed by AAAZZZS, e.g., 2350920.
elements (Elements) – A container for all the atomics elements information in the simulation.
nuclidesFile (str) – File path to the custom ARMI “nuclides.dat” file, containing a plain text description of all the nuclides to be modeled including: Z, number of neutrons, mass number, amu, natural abundance, half life and nu-bar and more.
mccNuclidesFile (str) – File path to the “mcc-nuclides.yaml” file, containing nuclides defined by the MC2-2 and MC2-3 codes, with various ENDF/B-V mappings.
- DEFAULT_NUCLIDES_FILE = '/home/runner/work/armi/armi/armi/resources/nuclides.dat'
- DEFAULT_MCC_NUCLIDES_FILE = '/home/runner/work/armi/armi/armi/resources/mcc-nuclides.yaml'
- factory(nuclidesFile: str = None, mccNuclidesFile: str = None, elementsFile: str = None)[source]
Reads data files to instantiate the
INuclides.Reads NIST, MC**2 and burn chain data files to instantiate the
INuclides. Also clears and fills in the class attibues: instances, byName, byLabel, byMcc3IdEndfbVII0, and byMcc3IdEndfbVII1. This method is automatically run upon initializing the class, hence it is not usually necessary to re-run it unless there is a change to the data files, which should not happen during run time, or a bad :py:class`INuclide` is created.- Parameters:
nuclidesFile (str) – File path to the custom ARMI “nuclides.dat” file, containing a plain text description of all nuclides to be modeled including: Z, number of neutrons, mass number, AMU, natural abundance, half life, nu-bar and more.
mccNuclidesFile (str) – File path to the “mcc-nuclides.yaml” file, containing nuclides defined by the MC2-2 and MC2-3 codes, with various ENDF/B-V mappings.
elementsFile (str) – File path to the custom ARMI “elements.dat” file.
Notes
This cannot be run more than once. NuclideBase instances are used throughout the ARMI ecosystem and are even class attributes in some cases. Re-instantiating them would orphan any existing ones and break everything.
- initReachableActiveNuclidesThroughBurnChain(nuclides, numberDensities, activeNuclides)[source]
March through the depletion chain and find all nuclides that can be reached by depleting nuclides passed in.
This limits depletion to the smallest set of nuclides that matters.
- Parameters:
nuclides (np.array, dtype="S6") – Starting array of nuclide names
numberDensities (np.array, dtype=np.float64) – Starting array of number densities
activeNuclides (OrderedSet) – Active nuclides defined on the reactor blueprints object. See: armi.reactor.blueprints.py
- isMonoIsotopicElement(name)[source]
Return true if this is the only naturally occurring isotope of its element.
- where(predicate)[source]
Return all
INuclidesobjects matching a condition.Returns an iterator of
INuclidesmatching the specified condition.- Parameters:
predicate (lambda) – A lambda, or function, accepting a
INuclideas a parameter
Examples
>>> from armi.nucDirectory.nuclideBases import NuclideBases >>> nuclideBases = NuclideBases() >>> [nn.name for nn in nuclideBases.where(lambda nb: "Z" in nb.name)] ['ZN64', 'ZN66', 'ZN67', 'ZN68', 'ZN70', 'ZR90', 'ZR91', 'ZR92', 'ZR94', 'ZR96', 'ZR93', 'ZR95', 'ZR'] >>> # in order to get length, convert to list >>> isomers90 = list(nuclideBases.where(lambda nb: nb.a == 95)) >>> len(isomers90) 3 >>> for iso in isomers: ... print(iso) <NuclideBase MO95: Z:42, A:95, S:0, label:MO2N> <NuclideBase NB95: Z:41, A:95, S:0, label:NB2N> <NuclideBase ZR95: Z:40, A:95, S:0, label:ZR2N>
- single(predicate)[source]
Return a single
INuclideobject meeting the specified condition.Similar to
where(), this function uses a lambda input to filter theINuclide instances. If there is not 1 and only 1 match for the specified condition, an exception is raised.Examples
>>> from armi.nucDirectory import nuclideBases >>> nuclideBases.single(lambda nb: nb.name == "C") <NaturalNuclideBase C: Z:6, w:12.0107358968, label:C> >>> nuclideBases.single(lambda nb: nb.z == 95 and nb.a == 242 and nb.state == 1) <NuclideBase AM242M: Z:95, A:242, S:1, label:AM4C>
- changeLabel(nuclideBase, newLabel)[source]
Updates a nuclide label and modifies the
byLabellook-up dictionary.Notes
Since nuclide objects are defined and stored globally, any change to the attributes will be maintained.
- imposeBurnChain(burnChainStream)[source]
Apply transmutation and decay information to each nuclide.
Notes
You cannot impose a burn chain twice. Doing so would require that you clean out the transmutations and decays from all the module-level nuclide bases, which generally requires that you rebuild them. But rebuilding those is not an option because some of them get set as class-level attributes and would be orphaned. If a need to change burn chains mid-run re-arises, then a better nuclideBase-level burnchain cleanup should be implemented so the objects don’t have to change identity.
See also
armi.nucDirectory.transmutationsdescribes file format
- addNuclideBases(nuclidesFile: str)[source]
Read natural abundances of any natural nuclides.
This adjusts already-existing NuclideBases and Elements with the new information.
This function reads the
nuclides.datfile from the ARMI resources folder. This file contains metadata for 4,614 nuclides, including number of protons, number of neutrons, atomic number, excited state, element symbol, atomic mass, natural abundance, half-life, and spontaneous fission yield. The data innuclides.dathave been collected from multiple different sources; the references are given in comments at the top of that file.- Parameters:
nuclidesFile (str) – File path to the custom ARMI “nuclides.dat” file, containing a plain text description of all nuclides to be modeled including: Z, number of neutrons, mass number, AMU, natural abundance, half life, nu-bar and more.
- readMCCNuclideData(mccNuclidesFile)[source]
Read in the label data for the MC2-2 and MC2-3 cross section codes to the nuclide bases.
This function reads the mcc-nuclides.yaml file from the ARMI resources folder. This file contains the MC2-2 ID (from ENDF/B-V.2) and MC2-3 ID (from ENDF/B-VII.0) for all nuclides in MC2. The
mcc2id,mcc3idEndfVII0, andmcc3idEndfVII1attributes of eachNuclideBaseinstance are updated as the data is read, and the global dictionariesbyMcc2IdbyMcc3IdEndfVII0andbyMcc3IdEndfVII1are populated with the nuclide bases keyed by their corresponding ID for each code.
- updateNuclideBasesForSpecialCases()[source]
Update the nuclide bases for special case name changes.
This function updates the keys for the
NuclideBaseinstances for Am-242m and Am-242 in thebyNameandbyDBNameglobal dictionaries. This function associates the more common isomer Am-242m with the name “AM242”, and uses “AM242G” to denote the ground state.Notes
This function is specifically added to change the definition of AM242 to refer to its metastable isomer, AM242M by default. AM242M is most common isomer of AM242 and is typically the desired isomer when being requested rather than than the ground state (i.e., S=0) of AM242.